
Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms
4.7
(214) ·
$ 4.50 ·
In stock
Description

Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins - Ritesh Gorkhali, Prashanna Koirala, Sadikshya Rijal, Ashmita Mainali, Adesh Baral, Hitesh Kumar Bhattarai, 2021

Plausible blockers of Spike RBD in SARS-CoV2—molecular design and underlying interaction dynamics from high-level structural descriptors
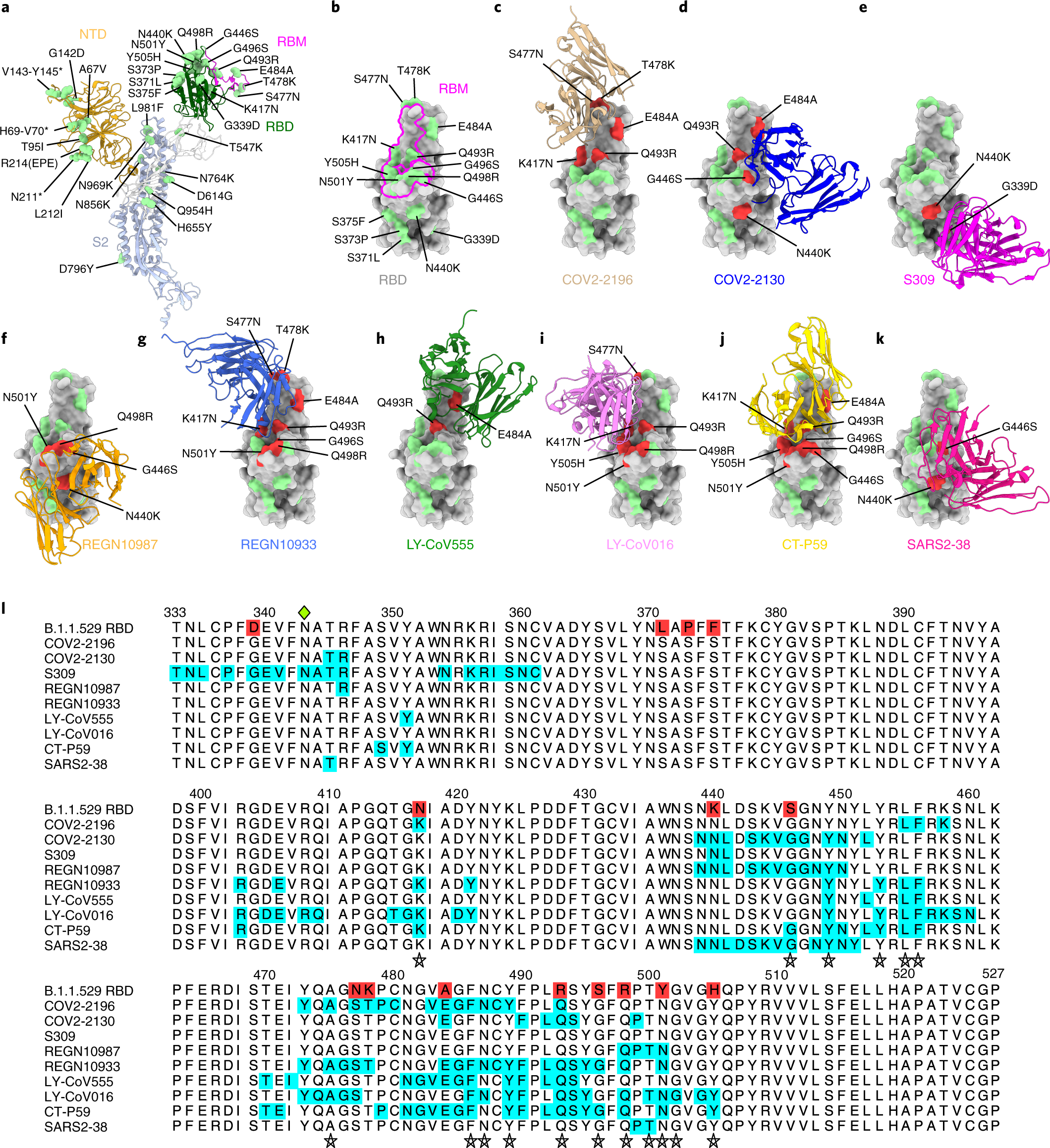
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies
Mutations in SARS-CoV-2 spike protein recepto

A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions

Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2

In-Silico Design of a Novel Tridecapeptide Targeting Spike Protein of SARS- CoV-2 Variants of Concern
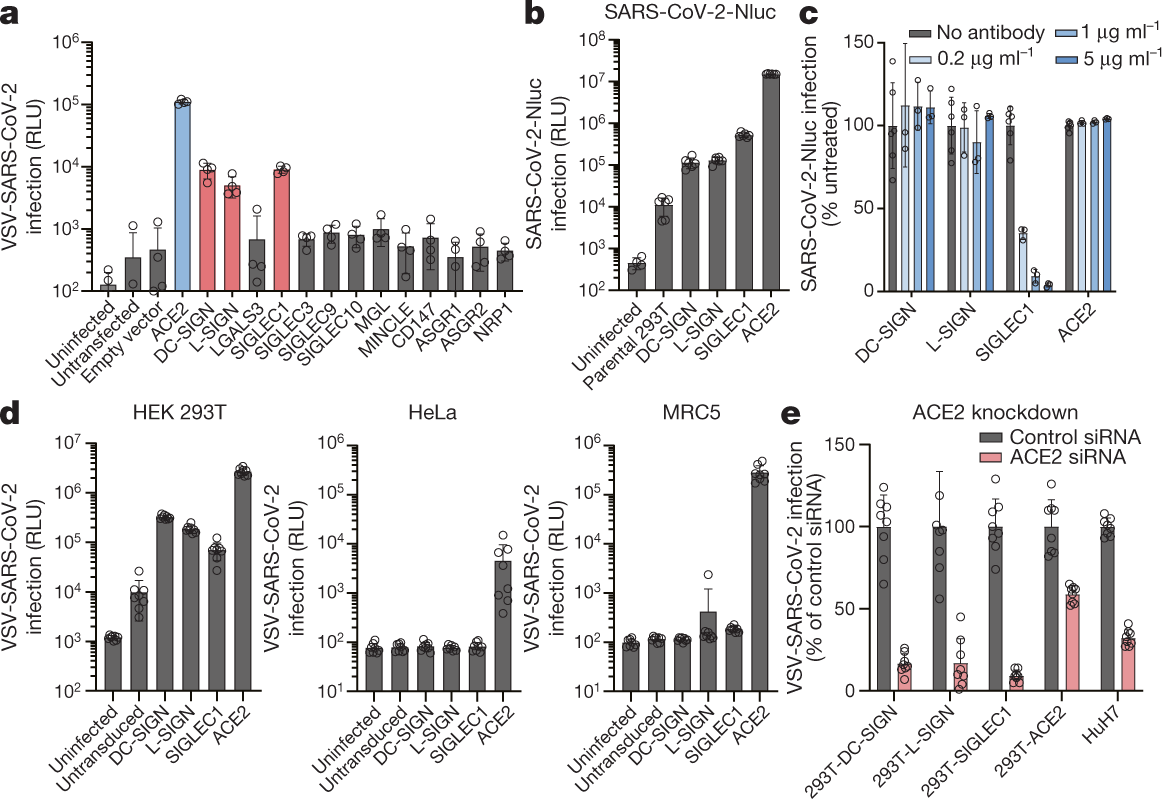
Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies

Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms
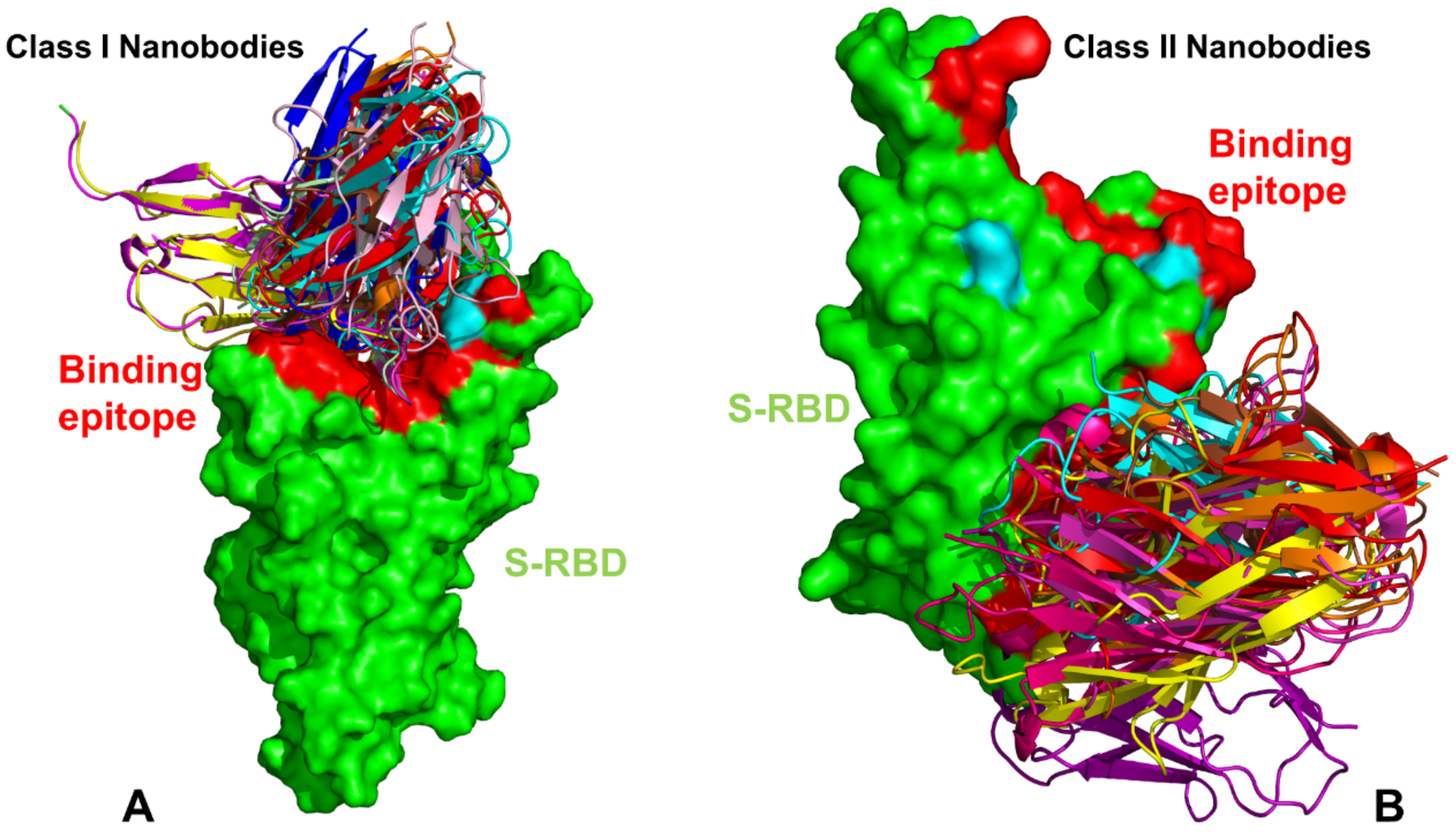
IJMS, Free Full-Text

Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions - ScienceDirect

Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2

Mutational landscape and in silico structure models of SARS-CoV-2 spike receptor binding domain reveal key molecular determinants for virus-host interaction, BMC Molecular and Cell Biology
Related products
You may also like
copyright © 2019-2024 transbytesystems.co.ke all rights reserved.








